DNA breaks in nerve cells' ancestors cluster in specific genes
The genome of developing brain cells harbors 27 clusters or hotspots where its DNA is much more likely to break in some places than others, researchers from the Program in Cellular and Molecular Medicine (PCMM) at Boston Children's Hospital, Harvard Medical School, and the Howard Hughes Medical Institute report in the journal Cell. Those hotspots appear in genes associated with brain tumors and a number of neurodevelopmental and neuropsychiatric conditions, raising new questions about these conditions' origins, as well as how the brain generates a diversity of circuitry during development.
The study's roots reach back more than 30 years, when PCMM director and study senior author Frederick Alt, PhD, and his colleagues first started investigating the links between tumors, oncogenes, DNA breaks and DNA repair, particularly in immune and nerve cells. Over the course of several studies, Alt's lab discovered that nerve cells lacking one particular DNA repair pathway called non-homologous end joining (NHEJ) -- which cannot repair breaks in their DNA strands -- either die off early in development or give rise to brain tumors called medulloblastomas.
However, they and other labs have struggled to understand why the loss of this pathway would have such dramatic effects.
"We've thought a lot about DNA breaks," said Alt, who is also a professor of genetics and the Charles A. Janeway Professor of Pediatrics at Harvard Medical School (HMS), "and many people over the years have considered the possibility that DNA breaks could be important for generating diversity in neural development. But nobody has had a way to identify the breaks in neural cells that would lead to this almost complete block of nervous system development in the absence of NHEJ."
In recent years, Alt's laboratory engineered a method for mapping, at very fine resolution, DNA breaks globally throughout the genome, called high-throughput genome-wide translocation sequencing (HTGTS). Initially developed to understand how genes reshuffle or translocate in cancer, the Alt lab has also used HTGTS to measure the precision of CRISPR gene editing and probe how the genome "sandboxes" DNA snipping enzymes to keep them from cutting genes in places they shouldn't.
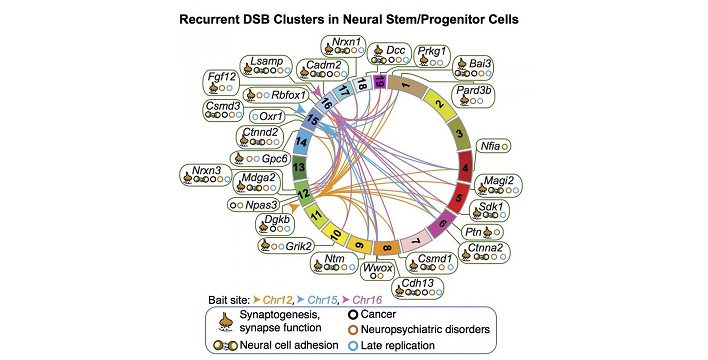
In the current study, Alt; lab members Pei-Chi Wei, PhD, Amelia Chang, Bjoern Schwer, MD, PhD; and their colleagues used HTGTS and informatics to search for and map DNA break patterns in mouse neural stem and progenitor cells (NSPCs, cells that produce the brain's neurons, astrocytes and oligodendrocytes) under conditions of replication stress.
The experiments revealed 27 clear, recurring hotspots where NSPCs' genomes break frequently. Strikingly, those 27 break hotspots were all spread across the bodies of 27 individual genes. Those genes share a number of characteristics in common:
- All were long, mostly more than 100 kilobases, with multiple exons (coding segments) and long introns (non-coding segments).
- Most are late replicators; that is, they are copied late in the cell division process.
- They encode proteins found on the surface of neurons that mostly perform functions that help neurons communicate (e.g., synapse formation, cell-cell adhesion).
- Twenty-four of the 27 genes have been linked to tumor suppression and/or neurological conditions such as autism spectrum disorder, schizophrenia and bipolar disorder.
"In our dreams we couldn't have found a set of genes to better fit the hypothesis that breaking DNA in neural cells is important," Alt exclaimed.
The breaks the team identified appeared most frequently in the gene's introns, leading the team to speculate that the hotspots might have a distinct purpose: to help the brain generate a diverse repertoire of circuitry. "Because the breaks occur mostly between exons, they would likely, in some cases, cause an exon or two to be deleted and potentially allow the gene to produce a different protein," Alt explained.
By splicing genes' exons in different ways, the genome could generate several variations of the proteins the genes encode. The neurons that develop from the NPSCs could then wire themselves into unique neural circuits.
"The protein encoded by one of the genes we identified, neurexin, potentially has more than 1,000 different forms, some of which may make connections between neurons of different strengths," said Wei, a postdoctoral fellow in Alt's laboratory. "What we found could provide a mechanism for making a diversity of synaptic connections and make contacts between neurons different."
"During neural development, you generate a whole brain, with some 100 billion neurons, from a relatively limited number of NSPCs," added Schwer, who is also an assistant professor of pediatrics at HMS. "In this context, is there a potential advantage to having recurrent DNA breaks? It could be a way to sample different combinations of circuits and synapses, almost like evolution in miniature.
"We don't know for sure that this is the case," he continued, "but we now show that these replication stress-associated breaks that occur during neural development could be a way to contribute to the perceived diversity of neural cells that end up in the mature brain."
The team also speculates, based on their findings, that replication stress-associated DNA damage during neural development could, by affecting these genes, promote neurodevelopmental or neuropsychiatric diseases.
"Virtually all of these genes have been associated with diseases that have a neurodevelopmental component," said Schwer. "It could be that when you can't efficiently repair breaks within genes, it could predispose the individual to neurodevelopmental disease."
Source: Boston Children's Hospital
BİOLOGY NEWS
-
RNA from Trained Aplysia Can Induce an Epigenetic Engram for Long-Term Sensitization in Untrained Aplysia.
-
HOW WE BECOME HAPPY
-
More tomatoes, faster: Accelerating tomato engineering
-
Purest yet liver-like cells generated from induced pluripotent stem cells
-
Legions of nanorobots target cancerous tumors with precision
-
CRISPR gene editing reveals new therapeutic approach for blood disorders
-
Quantum dots with impermeable shell: A powerful tool for nanoengineering
-
Thousands on one chip: New method to study proteins
-
Gene Drive Technology: Where is the future?
-
Four newly identified genes could improve rice
-
Watching the luminescent gene switch
-
DNA damage by ultrashort pulses of intense laser light
-
Mantis shrimp inspires next generation of ultra-strong materials
-
Specialized life forms abound at Arctic methane seeps
-
Ocean warming and acidification impact on calcareous phytoplankton